Performances of molecular-based assays for the quantification of viral genomes
Human Norovirus (NoV) and hepatitis A virus (HAV) are the main viruses suspected in foodborne outbreaks and represent a serious public health concern. These viruses are shed in large quantities in faeces and have a low infectious dose. Tools for the rapid detection of viral pathogens are essential for the analysis of clinical, environmental and food samples.
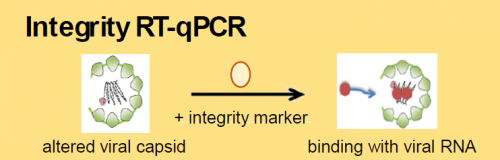
Currently, real-time RT-PCR is one of the most promising detection methods due to its sensitivity, specificity and speed. The EN ISO 15216 standard proposes molecular methods for the detection of NoV and HAV in high-risk food categories. Digital PCR (dPCR) technology has the potential to overcome some of the limitations of qPCR. It is an end-point, sensitive and accurate absolute quantification approach that enables the determination of target copy numbers without the need for external quantitative standards. Capsid integrity RT-qPCR, a molecular detection method for infectious viruses that combines integrity marker pre-treatment with RT-qPCR, has recently been developed in food virology for the selective detection of potentially infectious viral particles.
We tested the ISO 15216 method for virus extraction from alfalfa sprouts inoculated with NoV or HAV and a process control (murine norovirus, MNV). Virus recovery rates were high (ranging from 30% to 37 for all viral targets), with a very low PCR inhibition rate for each viral target (< 10% for all viral targets).
We then evaluated the performance of digital PCR and integrity RT-qPCR methods to quantify enteric viruses on spiked alfalfa sprouts. Virus recovery rates by digital RT-PCR (ranging from 34 to 36%) are comparable to those obtained by RT-qPCR for HAV and NoV. The quantification of HAV by integrity RT-qPCR is not affected by the presence of the alfalfa sprout matrix. This method discriminates between intact HAV particles and HAV particles with damaged capsids, with a 3.65 log10 reduction in genomic titer when the virus is inactivated at 80°C.
In conclusion, the molecular methods developed on alfalfa sprouts (RT-qPCR, RT-dPCR and integrity RT-qPCR) are suitable for the detection and quantification of enteric viruses and can be used to evaluate the sensitivity of each method and compare it with the performance of infectivity assays (HOLiFOOD WP2.7) and metagenomic assays (HOLiFOOD WP2.2).