Cellular based methods suitable for the detection of infectious virus
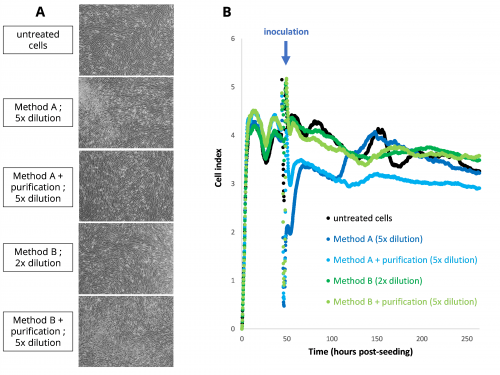
Morphology of FRhK-4 cells inoculated with alfalfa sprout eluates (A) and FRhK-4 cell index curves using the RTCA assay (B).
Human enteric viruses are a major public health concern because they can cause a range of acute illnesses, particularly acute gastrointestinal illness. These viruses can be transmitted through the consumption of contaminated water and food.
Although these viruses cannot replicate in the environment, they can survive for long periods on food, food contact surfaces and water. Among enteric viruses, human norovirus (NoV) and hepatitis virus (HAV) are the most important viruses suspected to cause foodborne outbreaks and represent a serious public health concern. Rapid detection of viral pathogens based on viral genome detection by molecular methods is critical for the analysis of clinical, environmental and food samples.
However, viral genome detection does not provide information on viral infectivity risk, and persistence data are essential for viral risk management. Detection of enteric virus infectivity is challenging due to the lack of a reliable cell culture method and the low levels of contamination in food and environmental samples. Murine norovirus (MNV) is a suitable surrogate for studying human norovirus infectivity because it is easy to propagate and titrate in cell culture and has structural and genetic properties that closely resemble those of human norovirus. Conventional cell culture-based methods such as the plaque assay or TCID50 can be used for inactivation studies and virus reduction analysis.
This report describes the achievement of Milestone M10, i.e., the performance of cell-based assays for the detection of infectious viruses on spiked alfalfa sprouts.
First, we evaluated different methods for the elution of HAV and MNV from alfalfa sprouts, and we observed the effect of the matrix on FRhK-4 cells (used for HAV infectious titration) and RAW 264.7 cells (used for MNV infectious titration) morphology using microscopy and viability using CCK-8 (Cell counting Kit-8) assay and RTCA (Real-Time Cell Analysis) assay. We determined the optimal dilution of the alfalfa sprout eluates (5x) at which no cytotoxic effect of the food matrix was observed. These elution methods were then evaluated for the detection and quantification of HAV and MNV spiked on alfalfa sprout eluates using the RTCA assay.
Alfalfa sprout eluates had no effect on the cell index profile of FRhK-4; whereas it affected the cell index profile of RAW 264.7 cells. However, virus titration can be performed for both RTCA assays because the correlation between the infectious virus titer of the inoculum and the tCI50 (time to 50% decrease in maximum cell index) is maintained in the presence of alfalfa sprout eluates, and the infectious virus titers are in the same range as the spiked virus titers. Moreover, the sensitivity of RTCA assays for the quantification of infectious HAV and MNV does not seem to be affected by the presence of alfalfa sprout matrix.
In conclusion, the virus titration methods developed to detect and quantify infectious viruses (HAV and MNV) on sprouted seeds can subsequently be used to evaluate the sensitivity of RTCA assays, and compare them with the performance of molecular-based assays and metagenomic assays. In addition, the developed RTCA assays will be used for viral persistence studies.




